Publications
Featured publications
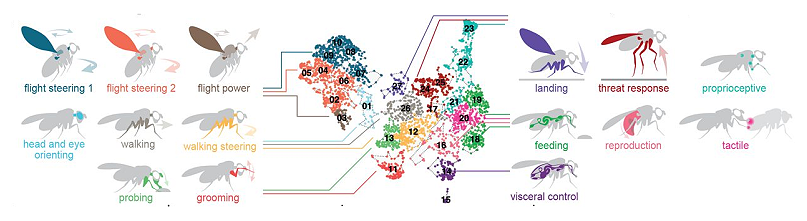
New! Distributed control circuits across a brain-and-cord connectome
bioRxiv preprint, Aug 2, 2025. DOI: 10.1101/2025.07.31.667571.
We are very excited to be part of the Brain-and-Nerve-Cord (BANC) project, a new resource which details the first complete connectome of an adult Drosophila melanogaster. This ground-breaking international effort represents the most complex full connectome of an adult animal to date, and offers intriguing new clues into the organisation of behavioural control across the nervous system.
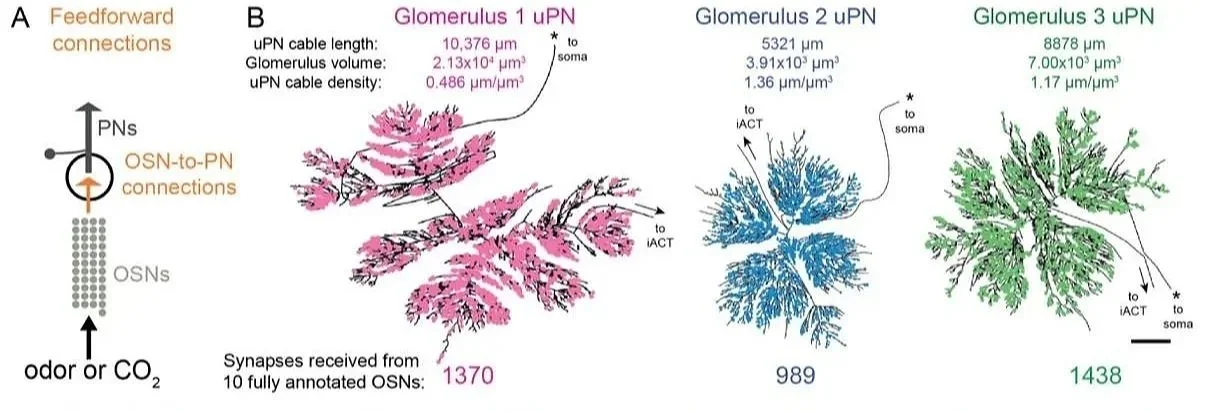
New! Recurrent connectivity supports carbon dioxide sensitivity in Aedes aegypti mosquitoes
bioRxiv preprint, July 30, 2025. DOI: 10.1101/2025.07.29.667487
Our image annotation and ground truthing services supported this fascinating project from the Lee lab at Harvard Medical School, which investigated the neural circuitry of carbon dioxide sensing in Aedes aegypti mosquitoes. This work reveals widespread recurrent connectivity within Aedes olfactory circuits, as well as ultrastructural findings including putative ribbon synapses, which have previously only been seen in vertebrates.
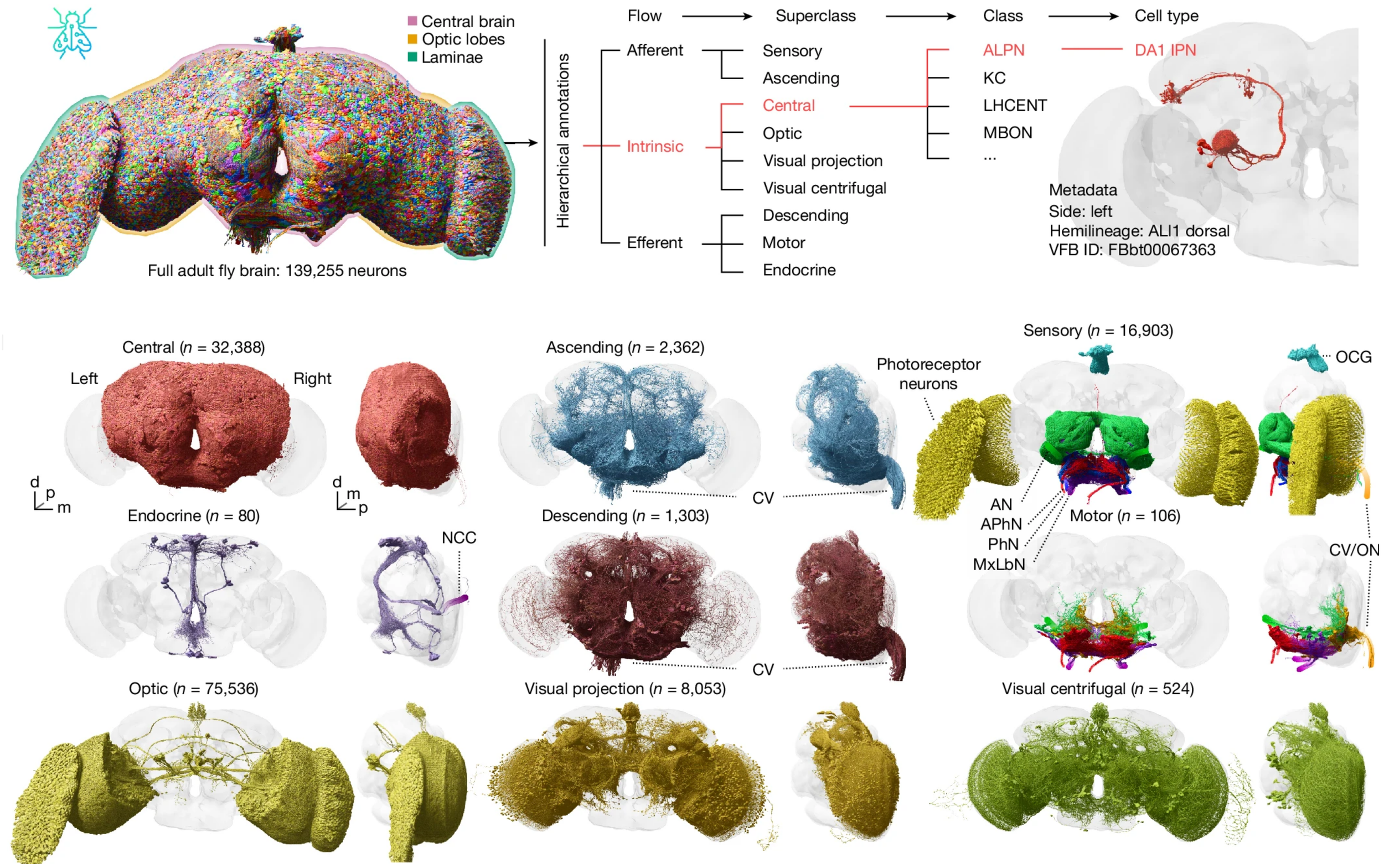
Whole-brain annotation and multi-connectome cell typing of Drosophila
Nature 364, Oct 2, 2024. DOI: 10.1038/s41586-024-07686-5
At Aelysia, we are proud to be part of the FlyWire Consortium, the international network of researchers behind the first complete adult brain map of the fruit fly Drosophila melanogaster. The FlyWire resource is described in detail by a collection of papers in Nature volume 364. Here, Schlegel et al. from the the MRC Laboratory of Molecular Biology describe a systematic, hierarchical annotation of all 140,000 neurons, as well as a new definition of cell typing based on intra- and inter-connectome comparisons.
All publications
Data annotation
J. Bao, W. Alford, A. Khandelwal, L. Walsh, G. Lantz, S. Poncio, L. Serratosa Capdevila, Y. Azatian, B. DePasquale, D.G.C. Hildebrand, M.A. Younger, W.C.A. Lee (2025) Recurrent connectivity supports carbon dioxide sensitivity in Aedes aegypti mosquitoes. bioRxiv. doi: https://doi.org/10.1101/2025.07.29.667487
S. Berg,*, I.R. Beckett, M. Costa, P. Schlegel, M. Januszewski, E.C. Marin, A. Nern, S. Preibisch, W. Qiu, S. Takemura, A.M.C. Fragniere, A.S. Champion, D-Y. Adjavon, M. Cook, M. Gkantia, K.J. Hayworth, G.B. Huang, W.T. Katz, F. Kämpf, Z. Lu, C. Ordish, T. Paterson, T. Stürner, E.T. Trautman, C.R. Whittle, L.E. Burnett, J. Hoeller, F. Li, F. Loesche, B.J. Morris, T. Pietzsch, M.W. Pleijzier, V. Silva, Y. Yin, I. Ali, G. Badalamente, A.S. Bates, J. Bogovic, P. Brooks, S. Cachero, B.S. Canino, B. Chaisrisawatsuk, J. Clements, A. Crowe1 , I.H. Vicente, G. Dempsey, E. Donà, M. dos Santos, M. Dreher, C.R. Dunne, K. Eichler, S. Finley-May, M.A. Flynn, I. Hameed, G. Patrick Hopkins, P.M. Hubbard, L. Kiassat, J. Kovalyak, S.A. Lauchie, M. Leonard, A. Lohff, K.D. Longden, C.A. Maldonado, M. Mitletton, I. Moitra, S. Soo Moon, C. Mooney, E.J. Munnelly, N. Okeoma1, D.J. Olbris, A. Pai, B. Patel, E.M. Phillips, S.M. Plaza, A. Richards, J. Rivas Salinas, R.J.V. Roberts, E.M. Rogers, A.L. Scott, L.A. Scuderi, P. Seenivasan, L. Serratosa Capdevila, C. Smith, R. Svirskas, S. Takemura, I. Tastekin, A. Thomson, L. Umayam, J.J. Walsh, H. Whittome, C.S. Xu, E.A. Yakal, T. Yang, A. Zhao, R. George, V. Jain, V. Jayaraman, W. Korff, G.W. Meissner, S. Romani1 , J. Funke, C. Knecht, S. Saalfeld, L.K. Scheffer, S. Waddell, G.M. Card, C. Ribeiro, M.B. Reiser, H.F. Hess, G.M. Rubin, G.S.X.E. Jefferis (2025) Sexual dimorphism in the complete connectome of the Drosophila male central nervous system. bioRxiv. https://doi.org/10.1101/2025.10.09.680999
A.S. Bates, J.S. Phelps, M. Kim, H.H. Yang, A. Matsliah, Z. Ajabi, E. Perlman, K.M. Delgado, M.A. Monium Osman, C.K. Salmon, J. Gager, B. Silverman, S. Renauld, M.F. Collie, J. Fan, D.A. Pacheco, Y. Zhao, J. Patel, W. Zhang, L. Serratosa Capdevila, R.J.V. Roberts, E.J. Munnelly, N. Griggs, H. Langley, B. Moya Llamas, R.T. Maloney, S. Yu, A.R. Sterling, M. Sorek, K. Kruk, N. Serafetinidis, S. Dhawan, T. Stürner, F. Klemm, P. Brooks, E. Lesser, J.M. Jones, S.E. Pierce-Lundgren, S. Lee, Y. Luo, A.P. Cook, T.H. McKim, E.C. Kophs, T. Falt, A.M. Negrón Morales, A. Burke, J. Hebditch, K.P. Willie, R. Willie, S. Popovych, N. Kemnitz, D. Ih, K. Lee, R. Lu, A. Halageri, J. A. Bae, B. Jourdan, G. Schwartzman, D.D. Demarest, E. Behnke, D. Bland, A. Kristiansen, J. Skelton, T. Stocks, D. Garner, F. Salman, K.C. Daly, A. Hernandez, S. Kumar; BANC-FlyWire Consortium; S. Dorkenwald, F. Collman, M.P. Suver, L.M. Fenk, M.J. Pankratz, G.S.X.E. Jefferis, K. Eichler, A.M. Seeds, S. Hampel, S. Agrawal, M. Zandawala, T. Macrina, D. Adjavon, J. Funke, J.C. Tuthill, A. Azevedo, H. S. Seung, B. L. de Bivort, M. Murthy, J. Drugowitsch, R.I. Wilson, W. A. Lee (2025) Distributed control circuits across a brain-and-cord connectome. bioRxiv. doi: https://doi.org/10.1101/2025.07.31.667571
S. Cachero, M. Mitletton, I.R. Beckett, E.C. Marin, L. Serratosa Capdevila, M. Gkantia, H. Lacin, G.S.X.E. Jefferis, E. Donà (2025) A Developmental Atlas of the Drosophila Nerve Cord Uncovers a Global Temporal Code for Neuronal Identity. bioRxiv. doi: https://doi.org/10.1101/2025.07.16.664682
T Stuerner*, P Brooks*, L Serratosa Capdevila, BJ Morris, A Javier, S Fang, M Gkantia, S Cachero, IR Beckett, P Schlegel, AS Champion, I Moitra, A Richards, F Klemm, L Kugel, S Namiki, HSJ Cheong, J Kovalyak, E Tenshaw, R Parekh, JS Phelps, B Mark, S Dorkenwald, AS Bates, A Matsliah, S Yu, CE McKellar, A Sterling, S Seung, M Murthy, J Tuthill, WA Lee, GM Card, M Costa, GSXE Jefferis, K Eichler (2025) Comparative connectomics of Drosophila descending and ascending neurons. Nature. 643, 158–172.
A Rayshubskiy, SL Holtz, A Bates, QX Vanderbeck, L Serratosa Capdevila, RI Wilson (2024) Neural circuit mechanisms for steering control in walking Drosophila. Elife, 13:RP102230.
P Schlegel, Y Yin, AS Bates, S Dorkenwald, K Eichler, P Brooks, DS Han, M Gkantia, M dos Santos, EJ Munnelly, G Badalamente, L Serratosa Capdevila, VA Sane, MW Pleijzier, IFM Tamimi, CR Dunne, I Salgarella, A Javier, S Fang, E Perlman, T Kazimiers, SR Jagannathan, A Matsliah, AR Sterling, S Yu, CE McKellar, FlyWire Consortium, M Costa, HS Seung, M Murthy, V Hartenstein, DD Bock, GSXE Jefferis (2024) Whole-brain annotation and multi-connectome cell typing quantifies circuit stereotypy in Drosophila. Nature, 634, 139–152.
S Dorkenwald, A Matsliah, AR Sterling, P Schlegel, S Yu, CE McKellar, A Lin, M Costa, K Eichler, Y Yin, W Silversmith, C Schneider-Mizell, CS Jordan, D Brittain, A Halageri, K Kuehner, O Ogedengbe, R Morey, J Gager, K Kruk, E Perlman, R Yang, D Deutsch, D Bland, M Sorek, R Lu, T Macrina, K Lee, JA Bae, S Mu, B Nehoran, E Mitchell, S Popovych, J Wu, Z Jia, M Castro, N Kemnitz, D Ih, A Shakeel Bates, N Eckstein, J Funke, F Collman, DD Bock, GSXE Jefferis, HS Seung, M Murthy, the FlyWire Consortium (2024) Neuronal wiring diagram of an adult brain. Nature, 634, 124–138.
MA Labouesse, M Wilhelm, Z Kagiampaki, AG Yee, R Denis, M Harada, A Gresch, A-M Marinescu, K Otomo, S Curreli, L Serratosa Capdevila, X Zhou, RB Cola, L Ravotto, C Glück, S Cherepanov, B Weber, X Zhou, J Katner, KA Svensson, T Fellin, L-E Trudeau, CP Ford, Y Sych, T Patriarchi (2024) A chemogenetic approach for dopamine imaging with tunable sensitivity. Nature Communications, 15, 5551.
HH Yang, LE Brezovec, L Serratosa Capdevila, QX Vanderbeck, A Adachi, RS Mann, RI Wilson (2024) Fine-grained descending control of steering in walking Drosophila. Cell, 187, 1–19.
M Harada, L Serratosa Capdevila, M Wilhelm, D Burdakov, T Patriarchi (2023) Stimulation of VTA dopamine inputs to LH upregulates orexin neuronal activity in a DRD2-dependent manner. Elife, 12:RP90158.
M Wilhelm, Y Sych, A Fomins, JL Alatorre Warren, C Lewis, L Serratosa Capdevila, R Boehringer, EA Amadei, B Grewe, EC O’Connor, BJ Hall, F Helmchen (2023) Striatum-projecting prefrontal cortex neurons support working memory maintenance. Nature Communications, 14, 7016.
P Schlegel*, AS Bates*, T Stürner, SR Jagannathan, N Drummond, J Hsu, L Serratosa Capdevila, A Javier, EC Marin, A Barth-Maron, IFM Tamimi, F Li, GM Rubin, SM Plaza, M Costa, GSXE Jefferis (2021). Information flow, cell types and stereotypy in a full olfactory connectome. eLife, 10:e66018.
AS Bates*, P Schlegel*, RJV Roberts, N Drummond, I Tamimi, R Turnbull, X Zhao, EC Marin, PD Popovici, S Dhawan, A Jamasb, A Javier, L Serratosa Capdevila, F Li, GM Rubin, S Waddell, DD Bock, M Costa, GSXE Jefferis (2020). Complete connectomic reconstruction of olfactory projection neurons in the fly brain. Current Biology, S0960-9822(20)30858-7.
Writing and editing
T Stuerner*, P Brooks*, L Serratosa Capdevila, BJ Morris, A Javier, S Fang, M Gkantia, S Cachero, IR Beckett, P Schlegel, AS Champion, I Moitra, A Richards, F Klemm, L Kugel, S Namiki, HSJ Cheong, J Kovalyak, E Tenshaw, R Parekh, JS Phelps, B Mark, S Dorkenwald, AS Bates, A Matsliah, S Yu, CE McKellar, A Sterling, S Seung, M Murthy, J Tuthill, WA Lee, GM Card, M Costa, GSXE Jefferis, K Eichler (2025) Comparative connectomics of Drosophila descending and ascending neurons. Nature.
P Schlegel, Y Yin, AS Bates, S Dorkenwald, K Eichler, P Brooks, DS Han, M Gkantia, M dos Santos, EJ Munnelly, G Badalamente, L Serratosa Capdevila, VA Sane, MW Pleijzier, IFM Tamimi, CR Dunne, I Salgarella, A Javier, S Fang, E Perlman, T Kazimiers, SR Jagannathan, A Matsliah, AR Sterling, S Yu, CE McKellar, FlyWire Consortium, M Costa, HS Seung, M Murthy, V Hartenstein, DD Bock, GSXE Jefferis (2024) Whole-brain annotation and multi-connectome cell typing quantifies circuit stereotypy in Drosophila. Nature, 634, 139–152.
N Eckstein*, AS Bates*, A Champion, M Du, Y Yin, P Schlegel, A Kun-Yang Lu, T Rymer, S Finley-May, T Paterson, R Parekh, S Dorkenwald, A Matsliah,7 S Yu, C McKellar, A Sterling, K Eichler, M Costa, S Seung, M Murthy, V Hartenstein, GSXE Jefferis, J Funke (2024) Neurotransmitter Classification from Electron Microscopy Images at Synaptic Sites in Drosophila Melanogaster. Cell, Vol. 34 (10), 2574-2594.
M Harada, L Serratosa Capdevila, M Wilhelm, D Burdakov, T Patriarchi (2023) Stimulation of VTA dopamine inputs to LH upregulates orexin neuronal activity in a DRD2-dependent manner. Elife, 12:RP90158.
M Wilhelm, Y Sych, A Fomins, JL Alatorre Warren, C Lewis, L Serratosa Capdevila, R Boehringer, EA Amadei, B Grewe, EC O’Connor, BJ Hall, F Helmchen (2023) Striatum-projecting prefrontal cortex neurons support working memory maintenance. Nature Communications, 14, 7016.
KR Mannion, EF Ballare, M Marks, T Gruber (2022) A multicomponent approach to studying cultural propensities during foraging in the wild. The Journal of Animal Ecology. DOI: 10.1111/1365-2656.13820. PMID: 36180982.
A Lazari, P Salvan, L Verhagen, M Cottaar, D Papp, OJ van der Werf, B Gavine, J Kolasinski, M Webster, CJ Stagg, MFS Rushworth, H Johansen-Berg (2022) A macroscopic link between interhemispheric tract myelination and cortico-cortical interactions during action reprogramming. Nature Communications, 13, 4253.
A Lazari, P Salvan, M Cottaar, D Papp, MFS Rushworth, H Johansen-Berg (2022) Hebbian activity-dependent plasticity in white matter. Cell Reports, Vol. 34 (11), 110951.
P Janz, MJ Nicolas, RL Redondo, M Valencia (2022) GABABR activation partially normalizes acute NMDAR hypofunction oscillatory abnormalities but fails to rescue sensory processing deficits. Journal of Neurochemistry, Vol. 1(5), 417-434.
E Parra-Barrero, K Diba, S Cheng (2021) Neuronal sequences during theta rely on behavior-dependent spatial maps. eLife, 10:e70296.
M Bainier, A Su, R L Redondo (2021) 3D printed rodent skin-skull-brain model: A novel animal-free approach for neurosurgical training. PLOS ONE, 16(6): e0253477.
EC Marin, L Büld, M Theiss, T Sarkissian, RJV Roberts, R Turnbull, IMF Tamimi, MW Pleijzier, WJ Laursen, N Drummond, P Schlegel, AS Bates, F Li, M Landgraf, M Costa, DD Bock, PA Garrity, GSXE Jefferis (2020). Connectomics analysis reveals first, second, and third order thermosensory and hygrosensory neurons in the adult Drosophila brain. Current Biology, S0960-9822(20)30844-7.
Contact us
We’re always excited to hear about new scientific projects. Contact us here and our team will get back to you soon.