FlyWire: the complete, annotated brain map of an adult Drosophila melanogaster
140,000 neurons, 50 million synapses. For the first time, the complete adult brain map of the fruit fly Drosophila melanogaster - one of biology’s most versatile model organisms - has been revealed in all its complexity [1, 2]. At Aelysia, we are proud to be part of the FlyWire Consortium, the international network of researchers behind one of the most ambitious connectomics projects ever realised.
In the field of connectomics, we seek to understand how nervous systems are wired. By painstakingly acquiring high-resolution brain images using electron microscopy, then manually and computationally reconstructing those images into a map, we can visualise neuronal morphology and connectivity down to the synaptic level. Until recently, this had only been possible for extremely small organisms such as Caenorhabditis elegans [3], which has a nervous system of only 302 neurons. In contrast, the D. melanogaster brain contains over a hundred thousand neurons, with an intricate anatomical and functional organisation that enables complex behaviour. Although the fly “hemibrain” connectome, published in 2020, represented a major step forward in our understanding of Drosophila neural circuitry [4], this partial reconstruction nonetheless left many questions unanswered - from how neuronal processes traverse more distal brain regions and inter-hemispheric connections, to the degree of stereotypy across hemispheres, and how representative a single connectome can be of morphology and connectivity across individuals.
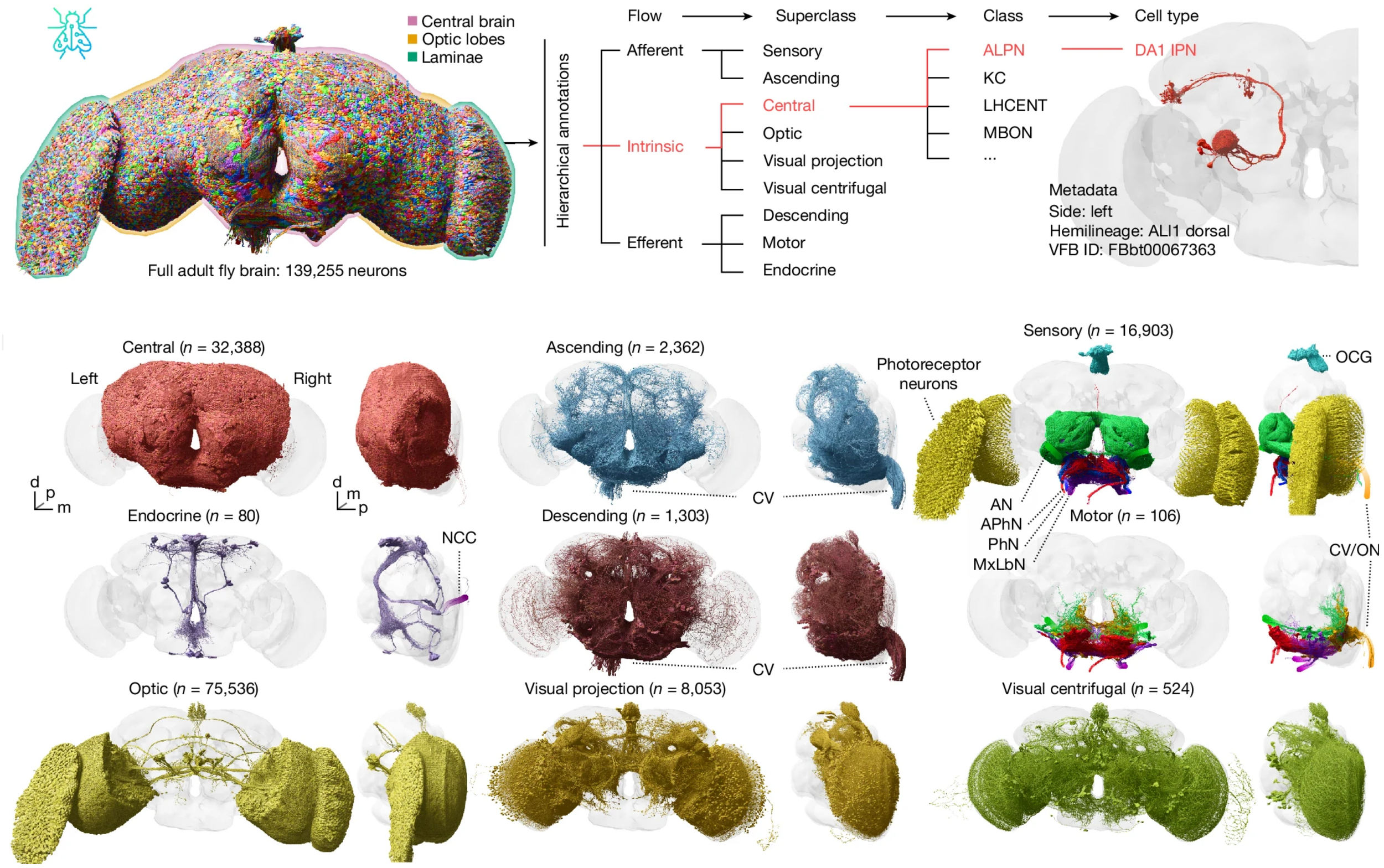
In a collection of articles published in Nature, the FlyWire Consortium now present a complete whole-brain connectome of an adult female D. melanogaster, comprising the central brain, optic lobes and suboesophageal zone, as well as partial reconstructions of the ascending and descending neurons which traverse the neck connective. Using a serial section transmission electron microscopy volume of the entire brain [5], generating the connectome relied on both automatic segmentation and on the collaborative efforts of expert and citizen science proofreaders, who contributed a staggering 33 person-years of manual proofreading. FlyWire comprises 139,255 neurons, of which 118,501 are intrinsic to the brain, and over 50 million chemical synapses. Furthermore, each neuron within the connectome has been comprehensively annotated for cell class and type, connectivity and predicted neurotransmitter identity [6], making this an invaluable, human-readable resource for Drosophila researchers worldwide.
In addition to the broad systematic grouping of cell classes and superclasses, FlyWire also includes a detailed annotation of neurons by developmental origin. During neuronal development, individual neuroblasts each divide into two hemilineages, with all daughter cells per hemilineage sharing a stereotyped morphology, co-fasciculation of neuronal processes within tracts, and fast neurotransmitter expression. Hemilineages therefore provide a natural functional and morphological grouping when studying the connectome. The FlyWire team successfully identified 120 neuroblast lineages and 183 hemilineages, accounting for 88% of all central brain neurons.
As a full brain reconstruction encompassing both hemispheres, FlyWire substantially advances our understanding based on the Drosophila hemibrain. First, FlyWire reveals the connectivity of bilateral- and contralateral-projecting neurons which were absent or truncated in the hemibrain, allowing crucial sensorimotor pathways to be investigated at the synaptic level. Second, as individual cell types in one hemisphere can be matched to those in the other, FlyWire shows that many types of neurons exhibit remarkable cross-hemispheric stereotypy. Third, by directly comparing FlyWire with the hemibrain, we can begin to understand how cell typing and connectivity patterns obtained in one connectome hold up to biological and technical variation - a key question in the connectomics field. Can we use the results of one connectome to validate those of another?
To answer this question, the FlyWire team sought to re-identify the over 5000 neuronal cell types originally proposed for the hemibrain connectome. After mapping hemibrain neurons into FlyWire space and matching neurons both manually and computationally, a majority of hemibrain cell types could be validated in FlyWire. However, even after splitting or merging individual cell types into composites, a substantial proportion (32%) of hemibrain cell types were simply not replicable across different animals. Instead, by taking advantage of cross-hemispheric and cross-brain comparisons, the FlyWire team propose a more robust, multiconnectome approach to cell typing. Here, a cell type is defined as a group of cells which are more similar to cells in another brain than to any cells in the same brain. Validating this approach across all three hemispheres showed that cell types and their connections are broadly stereotyped, with greater conservation amongst connections that met a threshold number of synapses. By reducing the complexity of the total connectome into functional units through cell typing, the FlyWire connectome can be used to inform network analyses and modelling, to generate hypotheses for functional experiments, and to understand biological differences via multiconnectomic approaches.
As part of the FlyWire Consortium, Aelysia supported the FlyWire connectome project through multiple rounds of expert proofreading, classification and seeding in different regions of the brain. Our work included completing and classifying antennal lobe neurons, seeding and editing visual projection neurons, and proofreading hemilineages for cross-brain comparisons. We also provided custom, detailed proofreading and cell type matching tasks in problem areas, fixing difficult segmentation errors, path swaps and artefact-related issues.
Click here to view the full collection of FlyWire articles published in Nature journals.
Explore the full, interactive FlyWire connectome at flywire.ai.
References:
Dorkenwald, S. et al. (2024): Neuronal wiring diagram of an adult brain. Nature doi:10.1038/s41586-024-07558-y
Schlegel, P. et al. (2024): Whole-brain annotation and multi-connectome cell typing of Drosophila. Nature doi:10.1038/s41586-024-07686-5
J.G. White et al. (1986): The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. doi:10.1098/rstb.1986.0056
Scheffer, L. K. et al. (2020): A connectome and analysis of the adult Drosophila central brain. eLife doi:10.7554/eLife.57443
Zheng, Z. et al. (2018): A complete electron microscopy volume of the brain of adult Drosophila melanogaster. Cell doi:10.1016/j.cell.2018.06.019
Eckstein, N. et al. (2024): Neurotransmitter classification from electron microscopy images at synaptic sites in Drosophila melanogaster. Cell doi:10.1016/j.cell.2024.03.016